
This gives rise to a bend in the helix axis. This is because the exposed C=O groups tend to point towards solvent to maximise their H-bonding capacity, ie tend to form H-bonds to solvent as well as N-H groups. Exposed helices are often bent away from the solvent region.


Proline occurs more commonly in extended regions of polypeptide. Helices containing proline are usually long perhaps because shorter helices would be destabilised by the presence of a proline residue too much. Janet Thornton has shown that proline causes two H-bonds in the helix to be broken since the NH group of the following residue is also prevented from forming a good hydrogen bond. This is because proline cannot form a regular alpha-helix due to steric hindranceĪrising from its cyclic side chain which also blocks the main chain N atom and chemically prevents it forming a hydrogen bond.
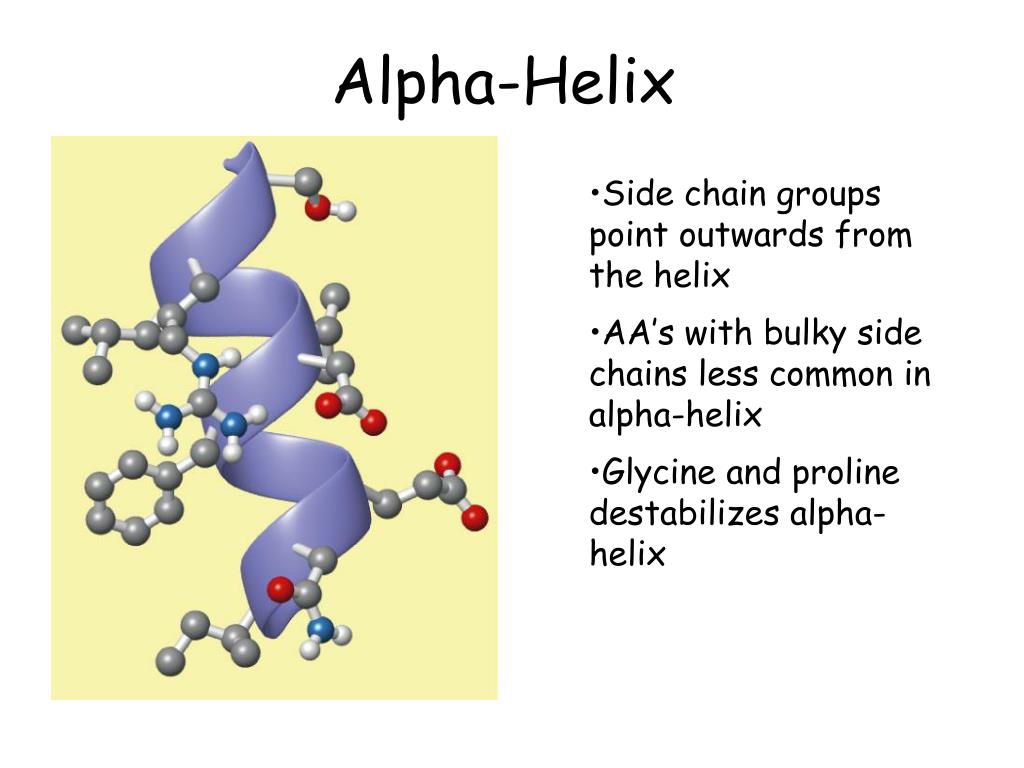
Proline residues induce distortions of around 20 degrees in the direction of the helix axis. The packing of buried helices against other secondary structure elements in the core of the protein. These distortions arise from several factors including: The majority of alpha-helices in globular proteins are curved or distorted somewhat compared with the standard Pauling-Corey model. All the amino acids have negative phi and psi angles, typical values being -60 degrees and -50 degrees, respectively. Side chains point outward from helix axis and are generally oriented towards its amino-terminal end. The peptide planes are roughly parallel with the helix axis and the dipoles within the helix are aligned, ie all C=O groups point in the same direction and all N-H groups point the other way. This gives a very regular, stable arrangement. Every mainchain C=O and N-H group is hydrogen-bonded to a peptide bond 4 residues away (ie O(i) to N(i+4)). The separation of residues along the helix axis is 5.4/3.6 or 1.5 Angstroms, ie the alpha-helix has a rise per residue of 1.5 Angstroms. Alpha-helices have 3.6 amino acid residues per turn, ie a helix 36 amino acids long would form 10 turns. The structure repeats itself every 5.4 Angstroms along the helix axis, ie we say that the alpha-helix has a pitch of 5.4 Angstroms. Bending is introduced by a strong (i, i + 8) hydrophobic interaction between the side chains of N-terminal tryptophan and leucine at the middle of the peptide sequence.Alpha-Helix Geometry Part. The peptide alpha-helix is bent despite the lack of an amphipathic sequence. The relative topologies of the charged side chains demonstrate flexibility and overall compromised favorable medium/long-range electrostatic interactions. The NMR structural ensemble demonstrates competition between E-R/K (i, i + 4) and (i, i - 1) ion pair formation, with the (i, i - 1) interactions being dominant. The presence of E-R/K (i, i + 4) ion pairs was expected to enhance the stability of the alpha-helix by introducing favorable electrostatic interactions at the side chain level, in addition to the characteristic backbone (i, i + 4) hydrogen bonds. The peptide sequence was designed using amino acids that have propensity for alpha-helix specificity. 
In addition, our data are consistent with the presence of several other transient and interconverting conformers. The peptide has a high population of a stable alpha-helical structure in the middle with fraying ends. We present the solution structure determination of a peptide with sequence Ac-WEAQAREALAKEAQARA-NH2, using NMR data.








 0 kommentar(er)
0 kommentar(er)
